
向 由起夫
(むかい・ゆきお)
Yukio Mukai
略歴
- 大阪大学大学院工学研究科博士後期課程修了
- 大阪大学工学部助手、同大学院工学研究科助手を経て、テキサス大学 M.D.アンダーソンがんセンターへ出張後、本学へ
応用微生物学研究室

卒業研究テーマ例
- ポリリン酸による細胞寿命制御メカニズムの解明
- 出芽酵母の機能未知遺伝子の機能解明
- 酒造に適した酵母の探索とその特性解析
細胞レベルの老化・寿命研究
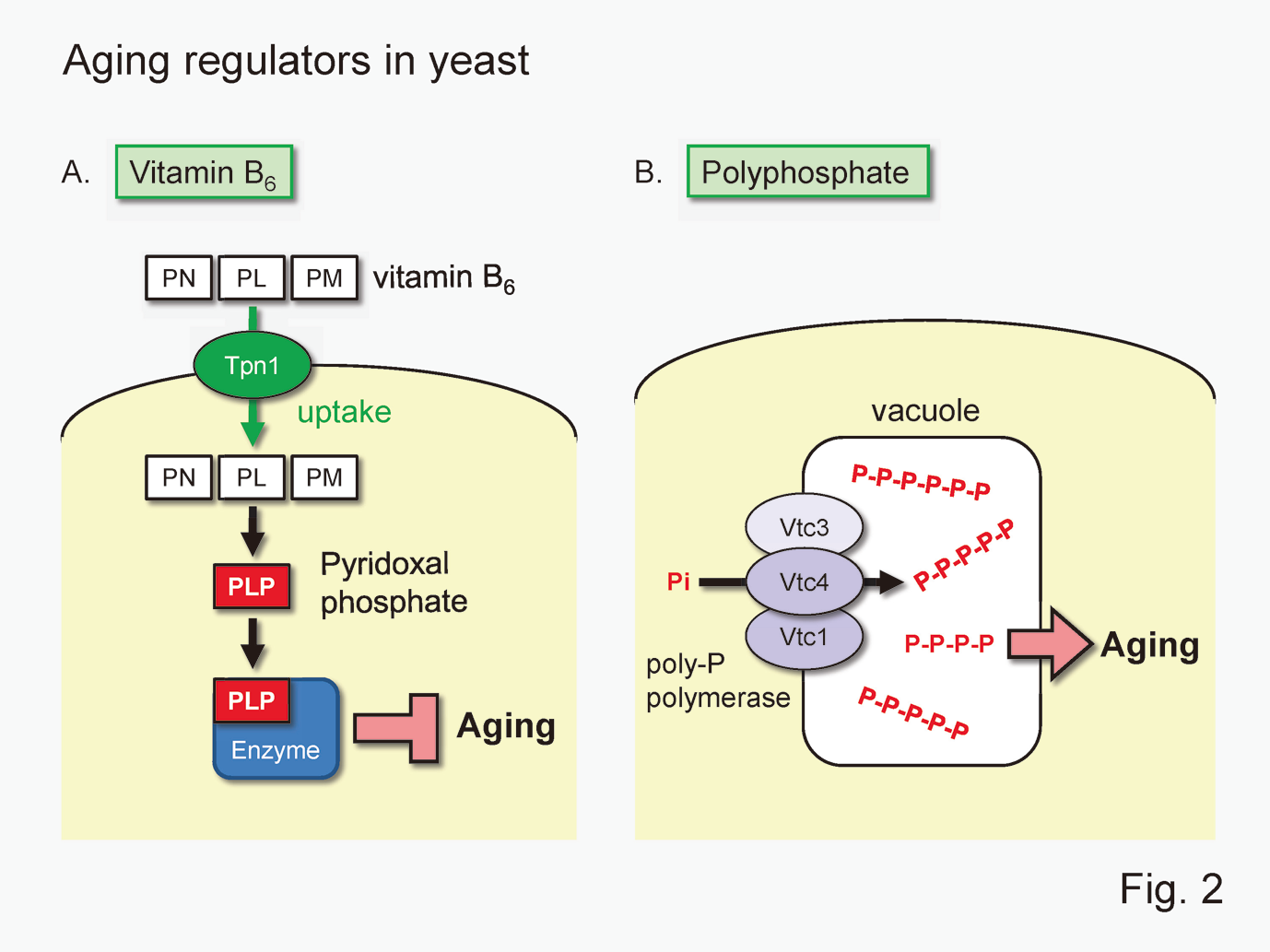
出芽酵母を用いた研究から、ビタミンB6が分裂寿命の維持に必要であり、ビタミンB6を必要とする代謝酵素の活性低下が細胞老化の原因になる可能性を示しました(Fig. 2A)。また、出芽酵母の細胞内にポリリン酸を蓄積させると分裂寿命が短くなることをみつけました(Fig. 2B)。今後、この酵母の知見をヒトの抗老化研究へ発展させていきます。
メタボロミクスによる遺伝子機能研究
- 研究の応用領域
- アンチエイジングの研究開発
実用酵母の育種
メタボロミクスによる遺伝子・細胞機能解析 - 産官学連携で求めるパートナー
- 老化・寿命研究に興味をもつ方
実用酵母を扱う企業
The aging society is progressing worldwide and elderly people (and young people) expect more healthy life years. Delaying the onset and progression of aging makes it possible to reduce the age-related diseases and prolong the life span. Hence, we are trying to discover the genetic and environmental aging factors and elucidate the mechanisms that control the aging process, for applying research to anti-aging. We also attempt to uncover the function of unknown genes via a metabolomics approach.
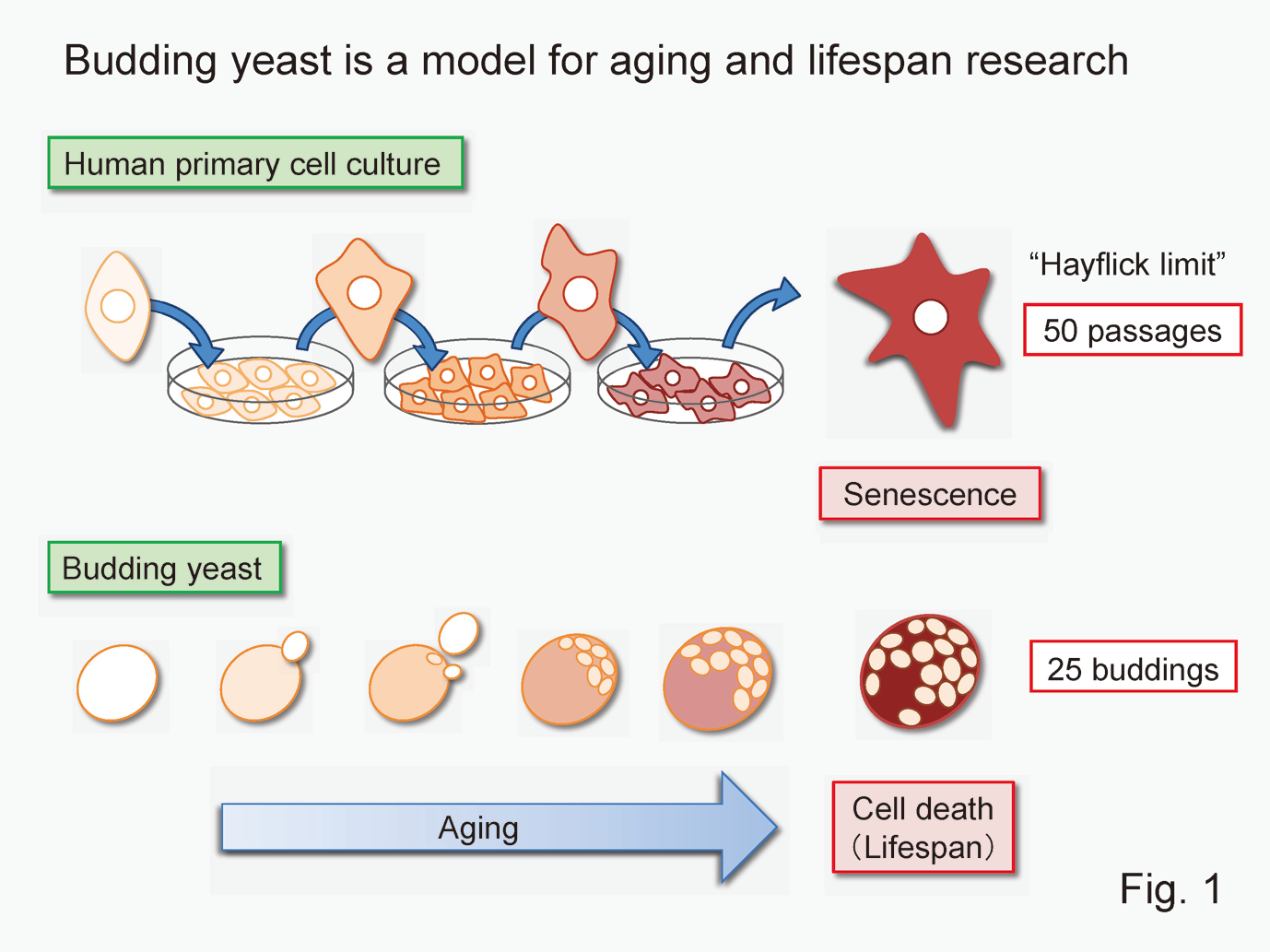
Cellular Aging and Lifespan Study
Cells as well as individuals have a finite lifespan, known as the Hayflick limit (Fig. 1). It is important to know how cellular lifespan is determined. The budding yeast Saccharomyces cerevisiae divides asymmetrically and has been investigated as a model organism for cellular aging and lifespan research. Yeast replicative lifespan is defined as the number of daughter cells produced by a mother cell before its senescence.
Recently, we showed that vitamin B6 is necessary to sustain the yeast replicative lifespan and suggested that decreases in vitamin B6-dependent enzyme activities cause cellular senescence (Fig. 2A). We also found that high accumulation of polyphosphate in yeast cells shortened the lifespan (Fig. 2B). These findings will be developed for human anti-aging research.


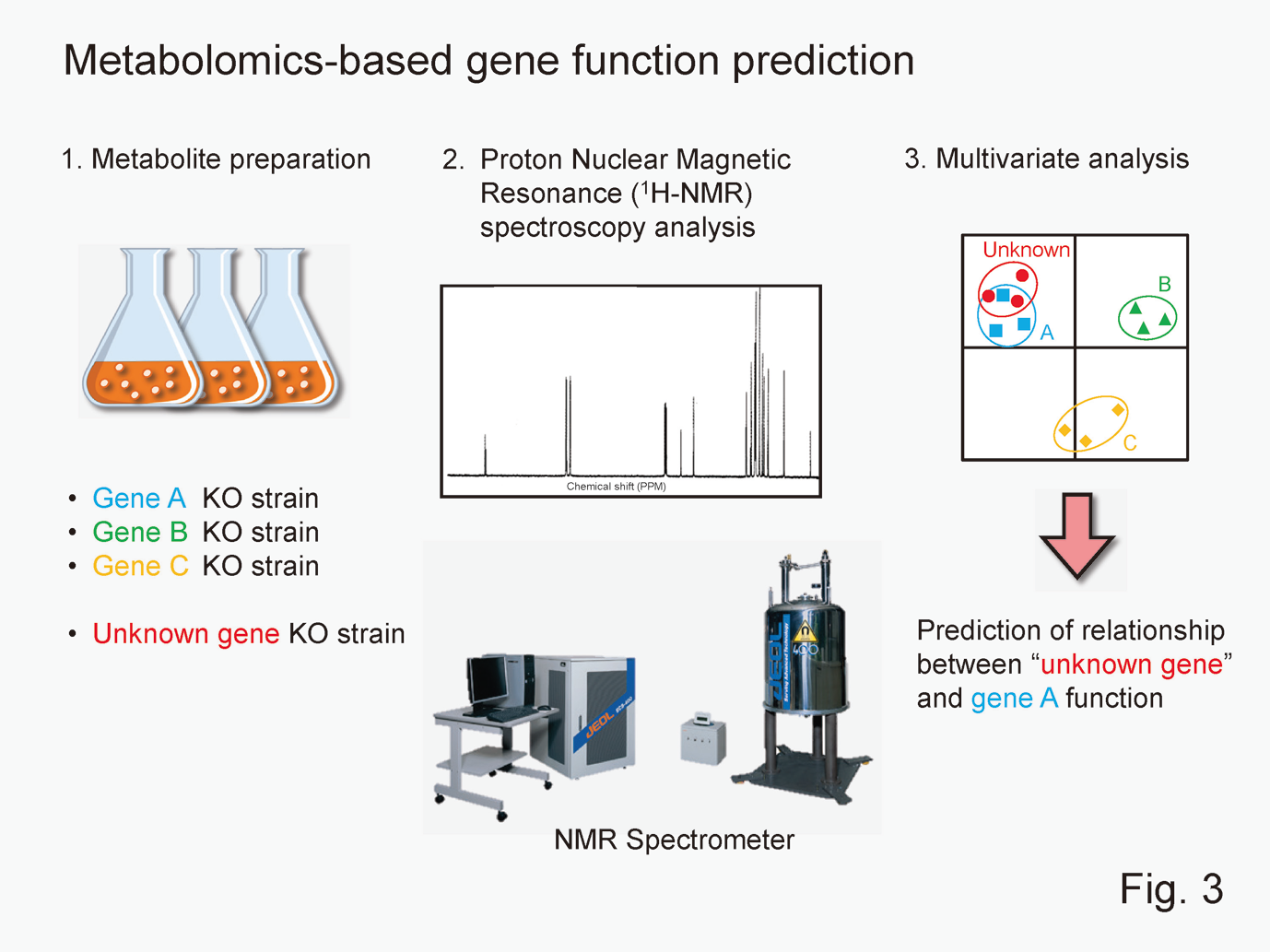
Metabolomics-based Gene Function Study

Among 6,600 protein-coding genes of the budding yeast, the function of about 700 genes remains unknown. Therefore, we obtain comprehensive information on metabolites of yeast strains disrupted for known and unknown genes by proton nuclear magnetic resonance spectroscopy method and predict the function of the unknown genes by comparing their metabolic profiling (Fig. 3). In addition, this metabolome analysis technique will be applied to the breeding of sake yeast for developing new sake.
Nakajima, T., S. Hosoyamada, T. Kobayashi, Y. Mukai. Secreted acid phosphatases maintain replicative lifespan via inositol polyphosphate metabolism in budding yeast. FEBS Lett. 596: 189-198. (2022)
Tai, A., Y. Kamei, Y. Mukai. The forkhead-like transcription factor (Fhl1p) maintains yeast replicative lifespan by regulating ribonucleotide reductase 1 (RNR1) gene transcription. Biochem. Biophys. Res. Commun. 488: 218-223. (2017)
Kamei, Y., A. Tai, S. Dakeyama, K. Yamamoto, Y. Inoue, Y. Kishimoto, H. Ohara, Y. Mukai. Transcription factor genes essential for cell proliferation and replicative lifespan in budding yeast. Biochem. Biophys. Res. Commun. 463: 351-356. (2015)
Kamei, Y., Y. Tamada, Y. Nakayama, E. Fukusaki, Y. Mukai. Changes in transcription and metabolism during the early stage of replicative cellular senescence in budding yeast. J. Biol. Chem. 289: 32081-32093. (2014)
Matsumura, H., N. Kusaka, T. Nakamura, N. Tanaka, K. Sagegami, K. Uegaki, T. Inoue, Y. Mukai. Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications. J. Biol. Chem. 287: 26528-26538. (2012)






