
塩生 真史
(しおにゅう・まさふみ)
Masafumi Shionyu
略歴
- 名古屋大学大学院理学研究科博士課程修了
- 名古屋大学大学院理学研究科助手を経て本学へ
生物情報解析学研究室

卒業研究テーマ例
- ランダムフォレストを使った金属イオン結合部位の予測
- 3D-Convolutional Neural Networkを用いたPLP結合部位予測
- Virtual Realityを用いたタンパク質の初期段階学習ソフトの開発
タンパク質と低分子化合物の結合予測
選択的スプライシングにより作られるタンパク質の機能性予測
- 研究の応用領域
- タンパク質のデザイン、創薬分野の技術開発
- 産官学連携で求めるパートナー
- 立体構造情報に基づいたタンパク質の機能解析を行いたい基礎・応用研究者、タンパク質工学に関連した企業、大学、国・地方自治体の研究機関
Thanks to advances in experimental technologies, many protein sequences and three-dimensional (3D) structures are accumulated in public databases. Based on these biological data, we are developing some bioinformatics methods for predicting protein functions / 3D structures of protein complexes. Moreover, to understand the characteristics of complicated protein 3D structures intuitively, we develop a visualization software of protein structures based on virtual reality.
Prediction of Interaction Between Proteins and Small Molecules

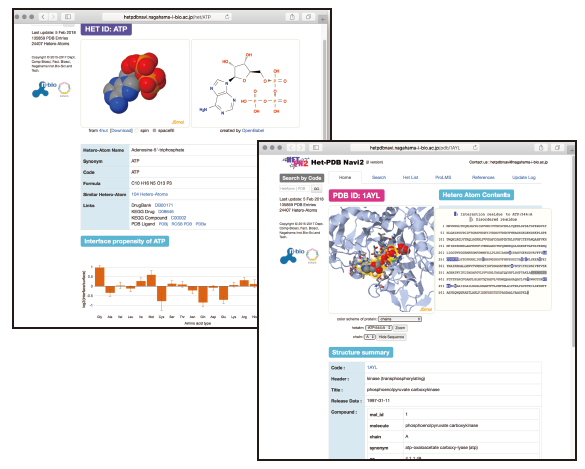
Known interface residues of proteins to small molecules, such as nucleotides, sugars, and ions, are valuable sources for predicting protein function and drug design. We provide information of interface residues to small molecules and statistics of them at Het-PDB Navi2 database(https://hetpdbnavi.nagahama-i-bio.ac.jp)(Fig.1). Furthermore, we are developing methods for predicting interface residues to small molecules based on the statistics obtained from Het-PDB Navi2 and machine learning algorithms.
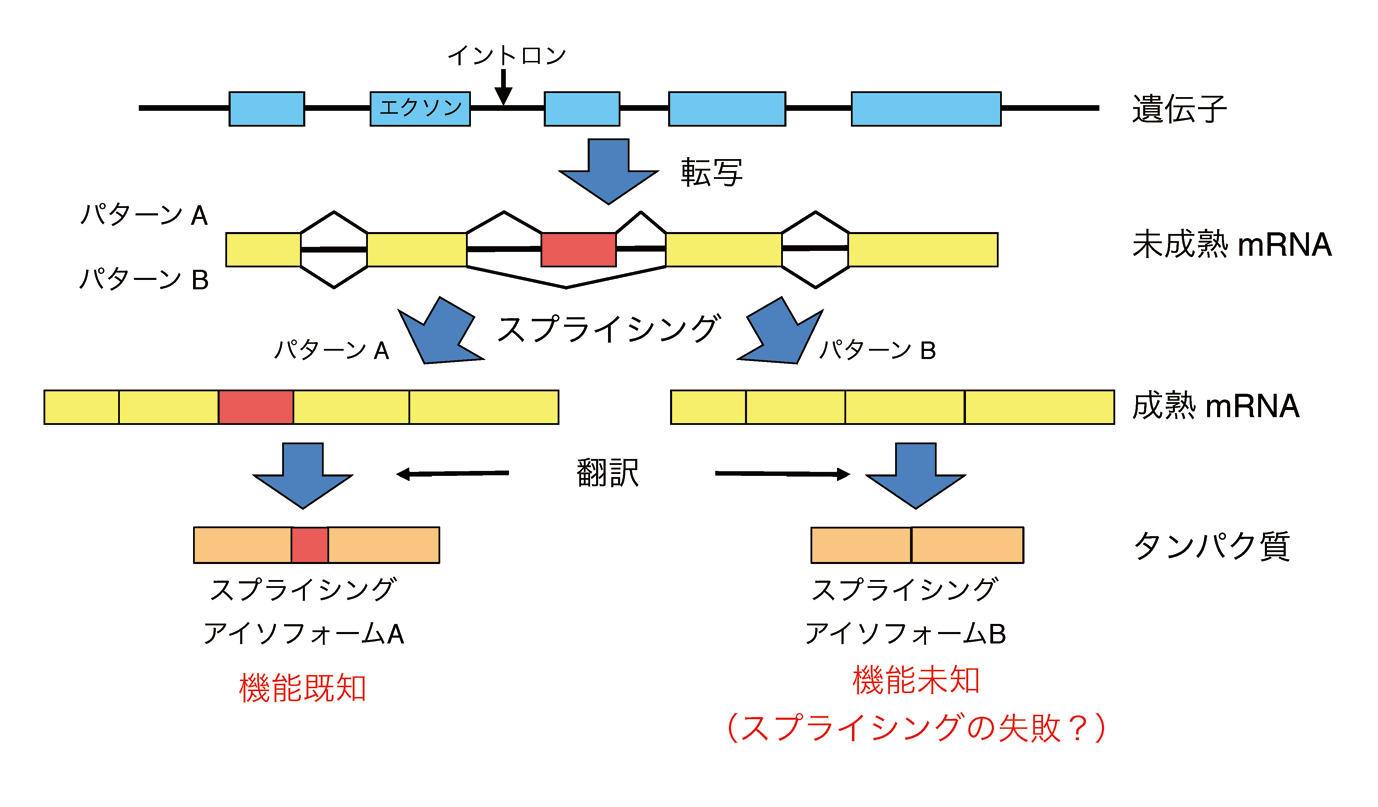
Development of Functionality Prediction Method for Splicing Isoforms
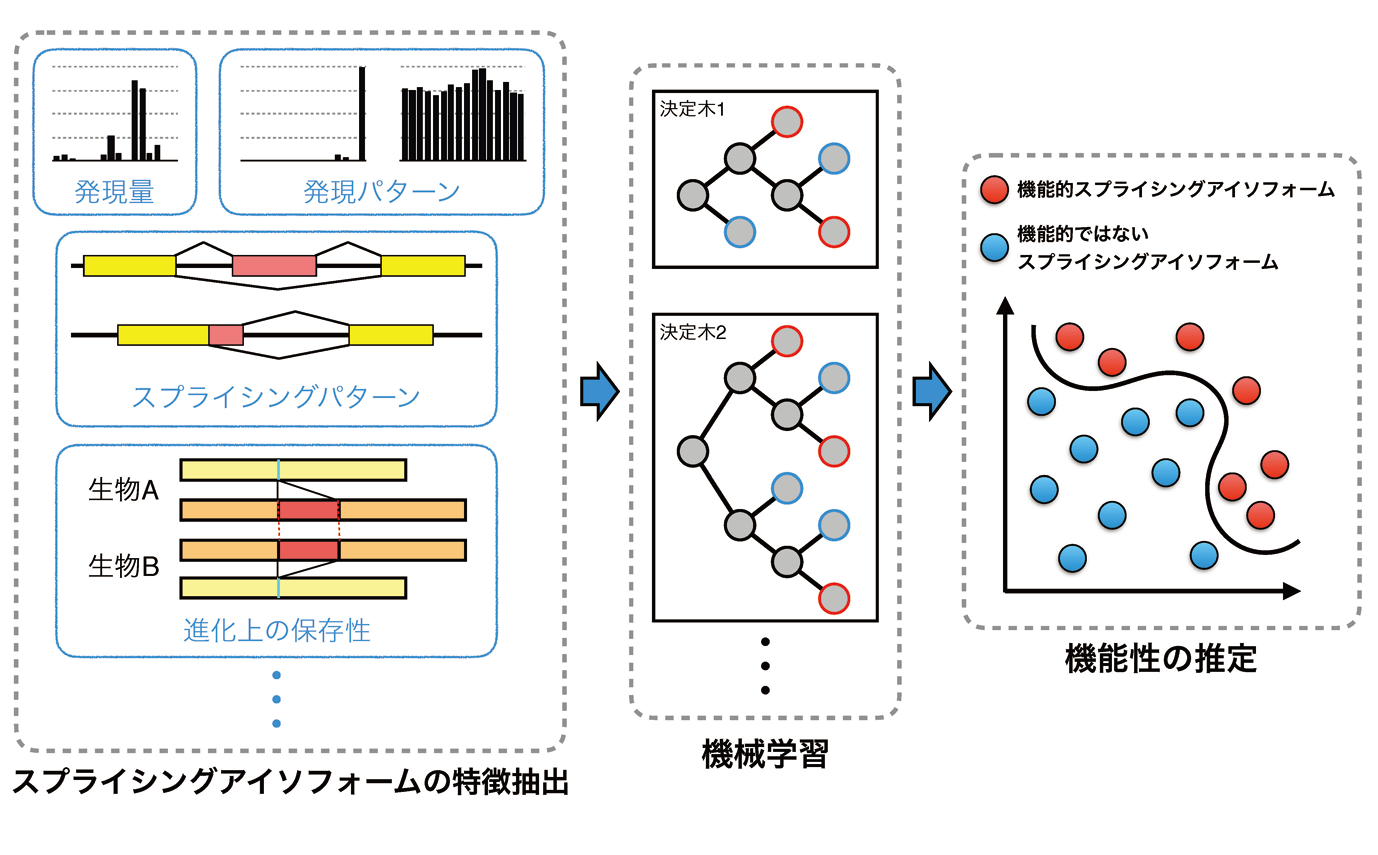
Alternative splicing is a molecular mechanism that produces multiple proteins from a single gene(Fig.2). Alternative splicing is thought to increase the number of proteins extensively over the number of genes in a genome and be required to generate complexity of higher eukaryotes, including humans. In many cases, however, functional differences among splicing isoforms at the molecular level are less known. Moreover, it is argued that alternative splicing may not contribute to proteome’s complexity because only a tiny number of splicing isoforms are confirmed to be expressed at the protein level by proteomics analyses. To clarify whether splicing isoforms are functional or not, we are developing a machine-learning-based algorithm that predicts the functionality of splicing isoforms based on the features of function-known splicing isoforms(Fig.3). We also provide the features of splicing isoforms used in this algorithm from the AS-ALPS database(https://as-alps.nagahama-i-bio.ac.jp)


Nakamae I, Morimoto T, Shima H, Shionyu M, Fujiki H, Yoneda-Kato N, Yokoyama T, Kanaya S, Kakiuchi K, Shirai T, Meiyanto E, Kato JY. Curcumin Derivatives Verify the Essentiality of ROS Upregulation in Tumor Suppression. Molecules, 24: 4067 (2019)
Tanaka M, Zhu Y, Shionyu M, Ota N, Shibata N, Watanabe C, Mizusawa A, Sasaki R, Mizukami T, Shiina I, Hasegawa M. Ridaifen-F conjugated with cellpenetrating peptides inhibits intracellular proteasome activities and induces drug-resistant cell death. Eur. J. Med. Chem., 146:636-650 (2018)
Shionyu M, Takahashi K, Go M, AS-EAST: a functional annotation tool for putative proteins encoded by alternatively spliced transcripts. Bioinformatics 28:2076-2077 (2012)
Shionyu M, Yamaguchi A, Shinoda K, Takahashi K, Go M, AS-ALPS: a database for analyzing the ef f ects of alternative splicing on protein structure, interaction and network in human and mouse. Nucleic Acids Res. 37(Database issue):D305-D309 (2009)
Yura K, Shionyu M, Hagino K, Hijikata A, Hirashima Y, Nakahara T, Eguchi T, Shinoda K, Yamaguchi A, Takahashi K, Itoh T, Imanishi T, Gojobori T, Go M. Alternative splicing in human transcriptome: functional and structural inf luence on proteins. Gene 380:63-71 (2006)






