
高橋 健一
(たかはし・けんいち)
Ken-ichi Takahashi
略歴
- 名古屋大学大学院理学研究科博士課程修了
- 名古屋大学大学院理学研究科助手を経て本学へ
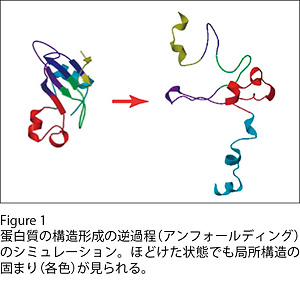
タンパク質のフォールディング過程のシミュレーション
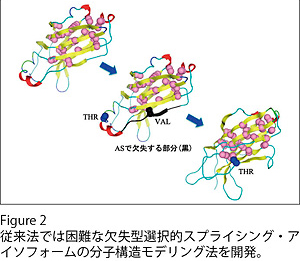
選択的スプライシング・アイソフォームの分子モデリング法の開発
生体分子の相互作用と構造変化の解析
- 研究の応用領域
- タンパク質・ゲノムの機能解析、タンパク質の設計
- 産官学連携で求めるパートナー
- 選択的スプライシングの関わる現象や疾病を研究対象とする企業、大学、国・地方自治体の研究機関
Protein and RNA are key molecular machines producing various biological phenomena. Through deep understanding of the physicochemical behavior of protein and RNA, fundamental mechanism of life must be clarified. In particular, toward understanding structural bases on which their functions are expressed, principles of self-organization of their native structures, and evolutionary processes producing such structures and functions, we have performed statistical analyses of structural data, molecular modeling and molecular simulation. Based on these studies, bioinformatics tools such as those for predicting structure and function of uncharacterized genes’ products and designing new useful molecules, will be also improved.
Molecular dynamics simulation of protein folding

Molecular mechanisms of self-organization to ordered native protein structures from disordered conformations have been studied by computer simulation of protein unfolding, a reverse process of the structural organization of proteins. In a stage of the unfolding process where a protein was extensively unfolded and kept on dynamically changing its overall conformation, we have found that local parts of the polypeptide chain tend to take relatively compact conformations. These parts correspond to compact substructures, “modules”, in the folded structure of the protein, which suggests a hierarchical process of protein folding (Figure 1). By utilizing this finding, we aim at accelerating in silico protein folding to find ordered native structures in practical time.
Development of molecular modeling method of alternative splicing isoforms

Alternative splicing (AS) is thought to be one of the major processes that increase the variation of product proteins sufficient to build complex molecular systems of eukaryotes. Structures and functions of most AS isoforms, however, remain to be elucidated. For inferring effects of AS on structures and functions of protein products and proposing experimental targets of AS isoforms, we have been developing a new strategy for molecular modeling of alternative splicing isoforms, especially for cases where large structural differences from template structures are anticipated because of the deletion of exons encoding parts of structural core regions (Figure 2).
Analysis of interaction and structural change of biomolecules
In approaching the subject of protein operating principle from mutant phenotype data, it is important to ask how the mutations affect protein structure and inter-molecular interaction. Specifically, how do mutations to proline within helices bend the helical structures and affect the binding surface to other molecules? Aiming to develop an estimation method for such phenomena, we have undertaken statistical analyses of proline-containing helices collected from the protein structure database, and also evaluated the reliability of molecular simulation as a method to investigate structural effects of mutations.
Shionyu M., Takahashi K. & Go M. AS-EAST: a functional annotation tool for putative proteins encoded by alternatively spliced transcripts. Bioinformatics 28:2076-2077 (2012)
Shionyu M., Yamaguchi A., Shinoda K., Takahashi K. & Go M. AS-ALPS: a database for analyzing the effects of alternative splicing on protein structure, interaction and network in human and mouse. Nucleic Acids Res. 37:D305-309 (2009)
Shinoda K., Takahashi K. & Go M. Retention of local conformational compactness in unfolding of barnase: contribution of end-to-end interactions within quasi-modules. BIOPHYSICS 3:1-12 (2007)
Takahashi K., Baba S., Koyanagi Y., Yamamoto N., Takaku H. & Kawai G. Two basic regions of NCp7 are sufficient for conformational conversion of HIV-1 dimerization initiation site from kissing-loop dimer to extended-duplex dimer. J. Biol. Chem. 276:31274-31278 (2001)
Takahashi K., Noguti T., Hojo H., Ohkubo T. & Go M. Conformational characterization of designed minibarnase. Biopolymers 58:260-267 (2001)






